Jumping genes, such as the ones that give this corn its distinctive coloration, have provided a new gene-editing mechanism.Credit: Getty
A molecular oddity found in bacteria could hold the key to redesigning genomes at will, allowing researchers to insert, delete or flip large segments of DNA.
The technique, described in three papers published this month in Nature1,2 and Nature Communications3, harnesses the natural ability of mobile genetic sequences, called jumping genes, to insert themselves into genomes.
Guided by an RNA molecule called a ‘bridge’ RNA or ‘seekRNA’, the system has been shown to edit genes in a bacterium and in test-tube reactions, but it is still unclear whether it can be adapted to work in human cells. If it can, it could be revolutionary, owing to its small size and its ability to make genetic changes that are thousands of bases long — much larger than is practical with the CRISPR–Cas9 genome-editing system — without breaking DNA.
“If this works in other cells, it will be game-changing,” says Sandro Fernandes Ataide, a structural biologist at the University of Sydney in Australia, and an author on the Nature Communications paper. “It’s opening a new field in gene editing.”
Transposable treasures
Like many celebrities, CRISPR–Cas9’s rise to fame has been plagued by misleading headlines. Although the method can be used to rewrite small segments of genomes, it is not the fully versatile cut-and-paste system that some news stories have made it out to be. The technique is most often used to change just one or a few DNA bases — and that typically requires first breaking the DNA and then relying on the cell’s innate DNA repair systems to generate the desired change. However, this opens the door to unintended collateral genetic damage as the cell implements the repair.
As CRISPR moves into human medicine, researchers are eager to expand their genome-editing toolkit so that they can insert entire or even multiple genes into a location of their choosing. Doing so would allow them to develop a therapy that treats people who have several mutations in a single gene, rather than targeting each mutation with a bespoke approach. And the ability to edit several genes could allow researchers to engineer immune cells to attack cancer in multiple ways, all while maintaining control over where those genes are inserted into the genome.
“What you really want to do in the future is to be able to design entire sections of your genome, not individual bases,” says Patrick Hsu, a bioengineer at the non-profit Arc Institute in Palo Alto, California, and an author of both of the Nature papers.
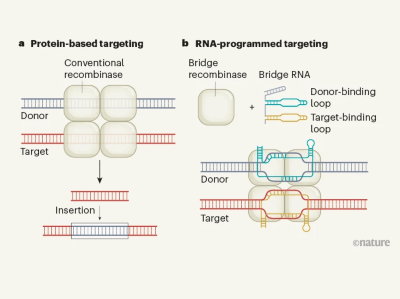
To look for tools, Hsu and his colleagues sifted through a diverse class of enzymes that allow mobile DNA elements in bacteria to hop from one location to another. They homed in on a family of transposable elements called IS110.
Enzymes in the IS110 family use a complex and unusual RNA-based targeting system, the team found. One end of the RNA binds to a piece of the DNA that will be inserted into the genome and the other end binds to a DNA snippet at the site in the genome where the cargo will go. Because the RNA bridges the two DNA segments, the team dubbed these molecules ‘bridge RNAs’.
By changing the sequences at either end of this bridge, the researchers were able to program IS110 enzymes to insert a cargo of their choosing where they wanted in the genome. They used the system to precisely insert a piece of DNA that was nearly 5,000 bases long into the genome of the bacterium Escherichia coli, and to excise and invert another piece of DNA from the E. coli genome.
Working independently of Hsu, Ataide and his colleagues characterized the biochemistry of IS110 molecules as well as those of another family, called IS1111, which uses a similar mechanism and is also programmable. They call their RNA intermediaries ‘seekRNA’.
Working out and exploiting these mechanisms is a remarkable achievement, says Elizabeth Kellogg, who studies mobile DNA elements called transposons at St. Jude Children’s Research Hospital in Memphis, Tennessee. “Everybody loves that transposons can insert large DNA cargoes,” she says, “but getting them to be programmable and site-specific is extremely difficult”.
Other transposition systems that researchers have explored for genome editing are more complex, she notes, and often consist of multiple proteins. In another paper published in Nature this month, researchers determined how key components of some of these elaborate machines form a complex structure known as a transpososome, which works together with an enzyme called transposase to allow mobile genetic elements to hop around in the genome4.
Size matters
Similarly, efforts to engineer CRISPR-based systems to make large manipulations in the genome often also require multiple proteins, or a fusion of a Cas enzyme with another protein. For example, a paper published on 26 June in Cell describes a method for duplicating chunks of the genome up to 100 million bases — larger than some human chromosomes — using a Cas9 protein joined to an enzyme that can copy the donor sequence5.
Programmable RNA-guided enzymes for next-generation genome editing
The IS110 and IS1111 systems, by contrast, require only a single protein, and this is less than half the size of many of the Cas enzymes used in CRISPR genome-editing systems. That size difference is important for medical applications: the viruses often used to ferry genome-editing components into human cells have limited cargo capacity.
But CRISPR systems also have the advantage of versatility, says Chengzu Long, a bioengineer at New York University Langone Health in New York City. Some Cas enzymes work in nearly every cell type studied.
The work on IS110 and IS1111 is “beautiful”, Long says. “But I really hope that in a few months, they’ll say it’s working in mice,” he adds. “Then, let’s have a cup of coffee.”
So far, IS110 family members do not seem to work well in mammalian cells, says Hiroshi Nishimasu, a structural biologist at the University of Tokyo who worked with Hsu to determine the mechanism by which an IS110 enzyme targets DNA. The team is now trying to engineer them to work better in the mammalian cells. Regardless of their success there, the IS110 mechanism stands out as a novel and “elegant” way by which mobile DNA elements can hitchhike around the genome, says Nancy Craig, a senior vice-president at SalioGen Therapeutics, a biotechnology company in Lexington, Massachusetts, that aims to develop genome-editing tools using mammalian transposons.
“Mother Nature has found many solutions for this,” she says. “We’ve found some, but there are many more out there waiting for us.”
Source link